Abstract
Background
The emergence and spread of carbapenem-resistant gram-negative bacteria pose a serious threat to human health. Currently, little is known about the molecular mechanisms underlying carbapenem -resistance and their prevalence among APEC in Egypt. The aim of this study was to detect APEC in clinically diseased broiler chickens collected from broilers farms located at Dakahalia governorates, asses their virulence –associated genes, detect the antimicrobial susceptibility of recovered isolates and to detect genes encoding carbapenemase resistant.
Methods
A total of 100 organ tissue samples subjected to conventional culture technique for isolation of E. coli. The confirmed E. coli were subjected to disc diffusion method for detection their susceptibility to antimicrobials. Polymerase chain reaction (PCR) was used for detection of APEC virulence genes (hlyA, iutA, ompT, iss, iroN) and six carbapenem- resistant genes namely, blaIMP, blaVIM, blaKPC, blaOXA-48 blaGES and blaNDM,.
Results
Forty isolates were confirmed to be E. coli among them, three or more APEC virulence- genes were detected from all isolates. The hlyA gene was detected in 90% (36/40), iroN in 95% (38/40), ompT in 97.5% (39/40), iutA in 92.5% (35/40) and iss was detected in 95% (38/40) of APEC isolates The tested isolates exhibited a remarkable resistance to ampicillin (97.5%), cefuroxime (92.5%), clindamycin (90%), chloramphenicol (62.5%), doxycycline (45%), amikacin (25%) and ciprofloxacin (12.5%). While, the retrieved isolates displayed 100 % sensitivity against imipenem, meropenem, ertapenem, ceftazidime and colistin. Concerning carbapenemase-encoding genes, blaIMP, blaVIM, blaKPC, blaOXA-48, blaGES couldn’t be detected among the E. coli isolates, while, blaNDM was confirmed in three isolates .
Conclusion
The detection of NDM as one of the carbapenem resistant genes reveals that the resistant strains are not only capable of infecting humans, but that carbapenams- resistant E. coli (CREC) has also started to pose a threat to poultry farm and other livestock animals. This may give rise to worries that these food-carrying creatures could infect humans or colonize them.
Author Contributions
Academic Editor: Mohammed A Elmetwally, Professor of Theriogenogy
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2023 Aya El- shaer, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Although E. coli is thought to be a normal component of the microflora in the chicken intestine, some strains can spread to other internal organs and result in lethal disease known as colibacillosis 1. Colibacillosis causes a huge economic losses in poultry farm. It caused by one of the most prevalent extraintestinal pathogenic (ExPEC), which cause infections outside of the gastrointestinal tract 2 known as Avian Pathogenic E. coli (APEC) 3, 4.
Multidrug resistance is a worrying problem that is being seen more frequently worldwide in both human and veterinary medicine 5. Although practically all therapeutically relevant antibiotics are generally ineffective against E. coli, this bacteria has a strong capacity to collect antibiotic resistance genes, primarily through horizontal gene transfer 6.
Carbapenems are considered the last-line antibiotics for treatment of microbial infection in human and it is commonly used to combat multiple resistant bacteria such as ESBL and MRSA which cannot be treated by other therapeutic options. However, there is concern that these carbapenemases will penetrate the food chain due to the recent discovery of this resistance in agricultural animals and poultry farms. Therefore, to maintain their effectiveness, the development and spread of resistance mechanisms against carbapenems have to be prevented. One benefit of this class of antibiotics is that carbapenems are comparatively resistant to hydrolysis by most -lactamases. They are however inactivated by carbapenemases, which also confer resistance to β-lactams 7. The genes coding for carbapenemases are frequently located in mobile genetic elements, facilitating their dissemination horizontally among different bacteria 8.
The appearance of carbapenemase-producing strains among gram-negative bacteria, particularly Enterobacteriaceae, has increased significantly over the past ten years, raising serious concerns and highlighting the need for prompt screening. Therefore, the aim of the present study was to detect presence of APEC in clinically diseased broiler chicken, detect the virulence- associated genes, investigate the antimicrobial susceptibility of recovered isolates and to identify the most common carbapenemase-encoding genes associated with the isolates under the study.
Material and methods
Samples collection
One- hundred unhealthy broiler chickens grown on commercial farms were included in this study. The selected birds were collected from commercial farm located in Dakahlia province, Egypt showed depression, high mortality rate, low body weight and loss of appetite. On postmortem examination, the birds showed pericarditis, fibrinopurulent, aerosaculitis and perihepatitis. The samples were transported under cold conditions to the laboratory in the Department of Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Mansoura University. All samples were processed within 3 hour after collection.
Isolation of Escherichia coli
Chicken organ samples were directly enriched in MaCconkey broth and incubated at 37°C for 18 hours. Then, a loopful from the overnight enriched broth culture was streaked onto Eosin Methylene Blue and MacConkey agar (Oxoid). The streaked plates were incubated at 37°C for 24 hours under aerobic condition. Suspected colonies (pink colored colonies on MacConkey agar and green colonies with metallic shin on EMB) were picked and purified on tryptic soya agar plates (TSA) and stored for further identification. Preliminary identification of E. coli was performed based on Gram staining and the other standard biochemical examination 9.
Antimicrobial susceptibility testing
Tests were carried out by the disk diffusion method on Mueller–Hinton agar and the results were interpreted according to the clinical breakpoints recommended by the Clinical and Laboratory Standards Institute 10. The following antibiotic discs (Oxoid, UK) were used: ampicillin (AMP; 10 µg), amikacin (AK; 30µg), cefuroxime (CXM; 30 µg), Colistin sulfate (CT, 10 µg), ceftazidime (CZN, 30 µg), imipenem (IPM, 10 μg), meropenem (MEM, 10 μg), ertapenem (ETP; 10 μg), ciprofloxacin (CIP; 5µg), clindamycin (DA; 2µg), chloramphenicol (C; 30µg (and doxycycline (DO; 30µg). Isolates classified as “intermediate” were grouped with “resistant” isolates. If one strain displayed resistance to ≥ 3 antimicrobial classes it defined as MDR.
Genomic DNA extraction
Genomic DNA were extracted by boiling of 3-5 colonies of suspected isolates for 10 min in 100 μl of DNA/RNA free water followed by centrifuged at 13,000 rpm for 10 minutes. The supernatant from boiled lysate was used as DNA template. The concentration of the obtained DNA were tested using a Nanodrop (Nanodrop 1000, Thermo Scientific, UK) 11.
Molecular characterization of E. coli
The conventional PCR was used to confirm the suspected E. coli isolates using 16S rRNA. The confirmed E. coli isolates were then subjected for PCR for detection of 5 virulence-associated genes including, hlyA, iutA, ompT, iss, iroN. The primers sets used for amplification were obtained from invetrogen/USA Table 1. PCR reaction and cycling condition were performed as previously described 12. In brief, 12.5 μL master mix (BioLab Inc., New England), 1 μL of forward and reverse primer of 10 pmol, 5.5 μL nuclease free water and about 5 μL DNA template was added to form a final volume of 25μL. The following thermal conditions was used: initial denaturation at 94 °C for 5 min, 35 cycles of 30 s at 94 °C, 1 min at 60 °C, and 1 min at 62 °C; the final extension was 72 °C for 5min. The PCR products were visualized by electrophoresis on 1.5% agarose gel in tris acetate buffer (TAE) and photographed.
Table 1. Oligonucleotide primers used in this study| Genes | Primer Sequence (5′–3′) | Amplicons(bp) | References |
| bla OXA-48 | Forward: GCGTGGTTAAGGATGAACAC Reverse: CATCAAGTTCAACCCAACCG | 438 | 15 |
| bla IMP | Foward:GGAATAGAGTGGCTTAAYTCTC Reverse: GGTTTAAYAAAACAACCACC | 232 | 15 |
| bla GES | Forward: AGTCGGCTAGACCGGAAAG Reverse: TTTGTCCGTGCTCAGGAT | 399 | 17 |
| bla VIM | Forward: GATGGTGTTTGGTCGCATAReverse: CGAATGCGCAGCACCAG | 390 | 15 |
| bla KPC | Forward: CGTCTAGTTCTGCTGTCTTGReverse: CTTGTCATCCTTGTTAGGCG | 798 | 15 |
| bla NDM-1 | Forward :GGTTTGGCGATCTGGTTTTCReverse :CGGAATGGCTCATCACGATC | 621 | 15 |
| 16 S rRNA | Foward :GACCTCGGTTTAGTTCACAGAReverse : CACACGCTGACGCTGACCA | 585 | 18 |
| iutA | F:GGCTGGACATCATGGGAACTGG R: CGTCGGGAACGGGTAGAATCG | 302 | 19 |
| iss | F:CAGCAACCCGAACCACTTGATG R: AGCATTGCCAGAGCGGCAGAA | 323 | 19 |
| ompT | F:TCATCCCGGAAGCCTCCCTCACTACTATR:TAGCGTTTGCTGCACTGGCTTCTGATAC | 496 | 19 |
| hlyA | F:GGCCACAGTCGTTTAGGGTGCTTACCR: GGCGGTTTAGGCATTCCGATACTCAG | 450 | 19 |
| iroN | F:AATCCGGCAAAGAGACGAACCGCCTR:GTTCGGGCAACCCCTGCTTTGACTTT | 553 | 19 |
Detection of carbapenem -resistance genes
Polymerase chain reaction (PCR) was performed to investigate the presence of carbapenemase-encoding genes, including blaKPC, blaNDM, blaVIM, blaIMP, blaGES and blaOXA-48 using PCR. The oligonucleotide primers (Invitrogen) are illustrated in Table 1 as previously reported 13, 14, 15, 16.The reaction was performed as mentioned above using the following thermal condition described in Table 2. PCR products were visualized in agarose gel electophorisis and using 1.5% agarose stained by ethidium bromide and photographed by the gel imaging system.
Table 2. Cyclic conditions used in PCR reactions.| Gene | Primary denaturation | Secondary denaturation | Annealing | Extension | No. of cycles | Final extension |
| 16S rRNA | 94˚C3 min. | 94˚C30 sec. | 60˚C1 min. | 68˚C2 min.. | 35 | 72˚C10 min. |
| bla NDM-1 | 94˚C5 min. | 94˚C30 sec. | 55˚C30 sec. | 72˚C30 sec. | 35 | 72˚C7 min. |
| bla GES | 95 ˚C15min | 95 ˚C1 min | 59 ˚C1min | 72 ˚C5 min | 30 | 72 ˚C |
| bla OXA-48 | 95 ˚C15 min | 95 ˚C1min | 60.5 ˚C1min | 72 ˚C5 min | 30 | 72 ˚C |
| bla IPM | 95 ˚C15 min | 95 ˚C1 min | 63 ˚C1 min | 72 ˚C5 min | 30 | 72 ˚C |
| bla VIM | 95 ˚C15 min | 95 ˚C1 min | 55 ˚C1 min | 72 ˚C5 min | 30 | 72 ˚C |
| bla KPC | 95 ˚C15 min | 95 ˚C1 min | 55 ˚C1 min | 72 ˚C5 min | 30 | 72 ˚C |
Please add space between E and coli
GES is written not in italic but only bla in italic
Please correct in al manuscript.
No results about virulence genes were reported here?
Abbreviation for what?
Results
Prevalence of E. coli in the tested samples
Organ samples were collected from clinically diseased chicken and were initially subjected to traditional methods of E. coli isolation. Among them, 40 E. coli isolates were recovered based on microscopical and biochemical identification. The suspected isolates were then confirmed by PCR assay targeting 16S rRNA (Figure 1).
Figure 1.Agarose gel electrophoresis showing amplification of hly gene (450bp) Lane 1-6, 9, 10 positive samples. Lane 7, 8, 11-14: negative samples.
Detection of carbabenemase encoding genes
PCR was used to identify carbapenemase genes or metallo-β-lactamase genes. Out of six gene which was subjected to PCR, NDM-1 gene was identified in 3 strains, while, blaIMP, blaVIM, blaKPC, blaOXA-48 and blaGES couldn’t be detected.
Distribution of virulence associated gene
Virulence -associated genes were screened by PCR assay using specific primers. The selected virulence- associated genes were detected in high prevalence rates. The hlyA gene was detected in 90% (36/40), iroN in 95% (38/40), ompT in 97.5% (39/40), iutA in 92.5% (35/40) and iss was detected in 95% (38/40) of APEC isolates (Figure 2, Figure 3, Figure 4, Figure 5).
Figure 2.Agarose gel electrophoresis showing amplification of hly gene (450bp) Lane 1-6, 9, 10 positive samples. Lane 7, 8, 11-14: negative samples.
Figure 3.Agarose gel electrophoresis showing amplification of iroN gene (553bp) Lane 1-6, 8: positive samples. Lane 7, 10-14: negative samples
Figure 4.Agarose gel electrophoresis showing amplification of iut Agene (302bp) Lane 1-6: positive samples. Lane 7 negative samples.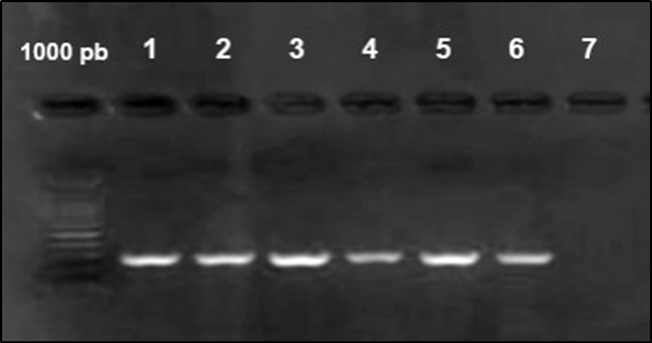
Figure 5.Agarose gel electrophoresis showing amplification of issgene (323bp) Lane 1-6: positive samples. Lane 7,8: negative samples.
Discussion
Colibacillosis is a common infectious disease caused by APEC, since that it results in significant financial losses for the poultry sector, this disease is a critical issue 17. This study aimed to detect APEC in broiler chicken symptomatic of colibacillosis as well as virulence-associated genes and carbapenem resistance genes among the retrieved APEC and profiling antimicrobial susceptibility in bacteria which are considered an important work to recognize the pathogenesis and possible hazards of anti-microbial resistance of APEC. In this study, 100 diseased broiler chickens `were collected from poultry farms and subjected for bacteriological examination, 40 (40%) E. coli isolates were recovered. Similarly, Hussein et al. 18 could isolate E. coli with a similar infection rate (43.1%) out of 800 chickens, while, Abd El Tawab et al. 19 reported a prevalence rate of 44% from imported chicken and 75% from local broiler chickens. A higher detection rate (48%) was also recorded 20. On the other hand, a lower rate was detected previously 21 with frequency of 34.95%.
Presence of virulence factors in bacterial cell increase their capacity to cause disease. The present findings consequently extend and corroborate that numerous putative virulence genes engage in the pathogenesis of colibacillosis. Since these genes were also discovered in colibacillosis isolates from various countries. It has been proposed that APEC retain these genes contributing to the development of colibacillosis. In this study, the virulence- associated genes selected were detected in high prevalence (90%, 95%, 97.5%, 92.5% and 95%) for hlyA, iroN, ompT, iut and iss respectively. The presence of these genes are associated with avian colibacillosis and indicates presence of APEC 22. The ompT may be involved in the pathogenesis of avian colibacillosis. It may also play a role in eukaryotic cell adhesion 23. In this study, ompT gene was detected in high percentage (97.5%) which is agreed with many previous reports detected this gene in high percentage among diseased chickens24, 25, 26, 27recorded a lower detection rate of this gene. Throughout the infection, the ompT promotes the formation of bacterial populations. Thus, this higher frequency indicated its important role APEC infection is indicated by its increased prevalence. Regarding iss gene, it is typically found on plasmids, produces a protein that contributes to serum resistance and complement resistance 28. Many previous reports could detected this gene in higher frequencies which are point to the possibility that this gene may play a key role in the development of avian colibacillosis. In this study, iss gene was identified in 95% of the retrieved E. coli isolates. Similarly, in Jordon, iss was identified in 93.3% of broiler chicken infected with collibacllosis 29. In the United States, iss gene was identified from 85.4% of APEC strains isolated from avian suffering from colibacillosis 30. As well as iss gene, was identified in a high prevalence rate (86.9%) from E .coli isolated from chickens with colibacillosis in Iran 31 . In this investigation, the frequency rate for the iutA gene was 92.5%. This outcome was consistent with a number of earlier international investigations as well 32, 33. However, Unno et al. 34 observed a significantly lower frequency (49%). The iroN is also detected in a high percentage in E. coli isolates which support previous reports that it may be important in the etiology of avian colibacillosis 35.
Infection with Carbapenem-Resistant Enterobacteriaceae (CRE) is emerging as an important challenge in health-care settings and a growing concern worldwide 36, 37. The incidence of carbapenem resistance in bacteria from animals hasn't received much attention, despite the fact that carbapenemases have been identified as a novel and perhaps developing concern in food-producing animals 38. The majority of epidemiological research to date has concentrated on human studies, with little work being done on animals used as food. In this study, a PCR was used for screening the retrieved E. coli isolates for the presence of carbabenem resistant genes. Among them, the NDM was identified in three isolates (7.5%; 3/40). Recently, a significant high frequency of NDM-1 has been also reported in China and India 39, 40. Additionally, a high prevalence rate of blaNDM (80%) was previously reported 41. The NDM was first discovered in Sweden in a patient who contracted an infection while travelling in India 42. Since then, the rapid spread of isolates that produce NDM via —MDR plasmids occurs raising the possibility that the widespread illnesses brought on by these strains will soon become incurable 43. Due to the widespread usage of antibiotics and the resulting high selection pressure, novel NDM-1 variants are emerging in India. Antibiotics for Gram-negative bacteria are scarce, and none of them are effective against NDM-1 producers 44. Other carbapenemase genes including blaIMP, blaVIM, blaKPC and blaOXA-48 were not detected in this study. Although carbapenems are rarely used to in in food, it's possible that the CREC evolved concurrently with resistance to other antimicrobials 45. The development of carbabenam resistance has been suggested that coselection of carbapenemase genes under the selection pressure imposed by the use of aminopenicillins and aminopenicillin—lactamase inhibitor combinations in farm animals 46
All obtained E. coli isolates were subjected to antimicrobial susceptibility test. In this study the highest resistant was recorded against ampicillin (97.5%), cefuroxime (92.5%), clindamycin (90%) and chloramphenicol (62.5%). While, 100% sensitivity was recorded against imipenem, meropenem and colistin and 87.5%, to ciprofloxacin, 75% to amikacin and 55% to doxycycline. Dissimilar to this study, Kumarasamy et al. 40 was reported complete resistance to rmeropenem and imipenem. While, Ho et al. 47 reported a resistance rates of 98.9 % (91/92), 91.3 % (84/92) and 95.7 % (88/92) to ertapenem, imipenem and meropenem. Similarly, in another study 7 E. coli isolates were resistant to ampicillin, cefotaxime, ceftazidime, cefepime, cefoxitin, ciprofloxacin, gentamicin, nalidixic acid, sulphamethoxazole, trimethoprim, meropenem, ertapenem, imipenen and, but a higher sensitivity was recorded against azithromycin, colistin and tetracycline 48. Carbapenem-resistant E. coli isolates were identified with carbapenems (including imipenem, meropenem, and ertapenem), MICs ranging from 2 μg/mL to ≧ 16 μg/mL 49. E. coli isolates were found to be resistant to imipenem and meropenem but lower resistance found in Hong Kong 50, 51, 52 in which carbapenem non-susceptible among clinical E. coli isolates remains rare and limited to sporadic occurrence.
Conclusion
Detection of carbapenem resistance genes, virulence-associated genes among APEC and profiling antimicrobial susceptibility in bacteria have been considered as an important work to recognize the pathogenesis and possible hazards of anti-microbial resistance of APEC. Colibacillosis can be prevented and controlled using antibiotics to treat the bacterial infections and to eliminate some predisposing causes. Therefore, restriction in antimicrobials use in poultry farms among veterinarians is highly recommended to control the spread of antimicrobial-resistant bacteria among poultry farm.
References
- 1.Ragione La, R M Woodward, J M.Virulence factors of Escherichia coli serotypes associated with avian colisepticemia. Avian Colibacillosis and Salmonellosis: A Closer Look at Epidemiology, Pathogenesis, Diagnosis, Control and Public Health Concerns , Res. Vet. Sci 2002, 27-35.
- 2.C D Köhler, Dobrindt U. (2011) What defines extraintestinal pathogenic Escherichia coli?. , International Journal of Medical Microbiology 301(8), 642-647.
- 3.L C Crossman, R, S A Beatson, T J Wells, Desvaux M et al. (2010) A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. , Journal of bacteriology 192(21), 5822-5831.
- 4.M A Croxen, B. (2010) Molecular mechanisms of Escherichia coli pathogenicity. , Nature Reviews Microbiology 8(1), 26-38.
- 5.Allocati N, Masulli M, M F Alexeyev, C Di Ilio. (2013) Escherichia coli in Europe: an overview. International journal of environmental research and public health. 10(12), 6235-6254.
- 6.Snyman Y, Whitelaw A C, Reuter S, MRB Maloba, Newton-Foot M.Colistin resistance mechanisms. in clinical Escherichia coli and Klebsiella spp. Isolates from the Western Cape of South Africa. Microb Drug Resist 27(9), 1249-1258.
- 7.Garcia-Graells C, Berbers B, Verhaegen B, Vanneste K, Marchal K et al. (2020) First detection of a plasmid located carbapenem resistant blaVIM-1 gene in isolated from meat products at retail in Belgium in 2015. International journal of food microbiology. 324-108624.
- 8.Monteiro J, R H Widen, A C Pignatari, Kubasek C, Silbert S. (2012) Rapid detection of carbapenemase genes by multiplex real-time PCR. , Journal of Antimicrobial Chemotherapy 67(4), 906-909.
- 9.Quinn P, Markey B, Carter M, Donnelly W, Leonard F. Veterinary Microbiology and Microbial Disease; Blackwell Science Company: Hoboken, NJ, USA; State University Press: (2002) . , Detroit, MI, USA
- 10.Clinical.Laboratory Standards Institute.2018. Performance standards for antimicrobial susceptibility testing. Twenty eight international supplement. Clinical and Laboratory Standards.
- 11.Ramadan H, Awad A, Ateya A. (2016) Detection of phenotypes, virulence genes and phylotypes of avian pathogenic and human diarrheagenic Escherichia coli in Egypt. , The Journal of Infection in Developing Countries 10(06), 584-591.
- 12.Amit-Romach E, Sklan D, Uni Z. (2004) Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poultry science. 83-7.
- 13.Mammeri H, Guillon H, Eb F, Nordmann P. (2010) Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC β-lactamases. Antimicrobial agents and chemotherapy. 54(11), 4556-4560.
- 14.Bokaeian M, Shahraki Zahedani S, Soltanian Bajgiran M, Ansari Moghaddam A. (2015) Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum beta-lactamases. , Jundishapur J. Microbiol 8, 10-5812.
- 15.E D Candan, Aksoz N. (2015) Klebsiella pneumoniae:characteristics of carbapenem resistance and virulence factors. , Acta Biochim. Pol 62, 867-874.
- 16.Sugawara E, Kojima S, Nikaido H. (2016) . Klebsiella pneumoniae Major Porins OmpK35 and OmpK36 allow more efficient diffusion of beta-lactams than their Escherichia coli homologs OmpF 198, 10-1128.
- 17.Dallenne C, Costa Da, Decré A, Favier D, C et al. (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. , Journal of Antimicrobial Chemotherapy 65(3), 490-495.
- 18.Amit-Romach E, Sklan D, Uni Z. (2004) Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poultry science. 83(7), 1093-8.
- 19.Ewers C, Janßen T, Kießling S, H C Philipp, L H Wieler. (2005) Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian diseases. 49(2), 269-273.
- 20.Chansiripornchai N. (2009) Comparative efficacy of enrofloxacin and oxytetracycline by different administration methods in broilers after experimental infection with avian pathogenic Escherichia coli. , The Thai Journal of Veterinary Medicine 39(3), 231-236.
- 21.Hussein H, Barakat H, M, Shoukry N. (2017) Endotoxin production from Escherichia coli O157 locally isolated from some Egyptian foods and its effect on liver efficiency in rats. , Journal of Environmental Science 40(3), 23-36.
- 22.Abd El Tawab, A, F I El-Hofy, M E El-khayat, H B Mahmoud. (2016) Prevalence of blaTEM and blaSHV genes in genomic and plasmid DNA of ESBL producing Escherichia coli clinical isolates from chicken. , Benha Veterinary Medical Journal 31(1), 167-177.
- 24.Momtaz H, Jamshidi A. (2013) Shiga toxin-producing Escherichia coli isolated from chicken meat in Iran: Serogroups, virulence factors, and antimicrobial resistance properties. , Poultry science 92(5), 1305-1313.
- 25.Timothy S, Shafi K, Leatherbarrow A H, FTW Jordan, Wigley P.Molecular epidemiology of a reproductive tract associated colibacillosis outbreak in a layer breeder flock associated with atypical avian pathogenic Escherichia coli. , Avian Pathol 37(4), 375-8.
- 26.H M, Zhu Y, Ma J, Zhang Y, Pan Z et al. (2017) Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. , Microbial pathogenesis 107, 29-37.
- 27.de Oliveira AL, Rocha D A, Finkler F, de Moraes LB, Barbieri N L et al. (2015) . Prevalence of ColV Plasmid- Linked Genes and In Vivo Pathogenicity of Avian Strains of Escherichia coli, Foodborne Path Dis: 12, 679-684.
- 28.Chalmers G, A C Cormier, Nadeau M, Côté G, R J et al. (2017) Determinants of virulence and of resistance to ceftiofur, gentamicin, and spectinomycin in clinical Escherichia coli from broiler chickens in. , Québec, Canada 203-149.
- 29.Varga C, Brash M L, Slavic D, Boerlin P, Ouckama R et al. (2018) Evaluating virulence-associatedg and antimicrobial resistance of avian pathogenic Escherichia coli isolates from droiler and broiler Breeder chickens in Ontario. , Canada. Avian Dis 62, 291-299.
- 30.Mbanga J, Nyararai Y O. (2015) Virulence gene profiles of avian pathogenic Escherichia coli isolated from chickens with colibacillosis in Bulawayo, Zimbabwe. , Onderstepoort J Vet Res 82(1), 850.
- 31.Binns M, Davies D L, Hardy K G. (1979) Cloned fragments of the plasmid ColV, I-K94 specifying virulence and serum resistance. , Nature 279(5716), 778-781.
- 32.R A Ibrahim, T L Cryer, S Q Lafi. (2019) Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. , BMC Vet Res 15, 159.
- 33.Dissanayake D R, Octavia S, Lan R. (2014) Population structure and virulence content of avian pathogenic Escherichia coli isolated from outbreaks in Sri Lanka. , Vet Microbiol 168, 403-412.
- 34.Bonjar S, Salari S, Jahantigh M, Rashki A. (2017) Frequency of iss and irp2 genes by PCR method in Escherichia coli isolated from poultry with colibacillosis in comparison with healthy chicken in poultry farms of Zabol, South East of Iran. Polish journal of veterinary sciences.
- 35.Schouler C, Schaeffer B, Bree A, Mora A, Dahbi G et al. (2012) Diagnostic strategy for identifying avian pathogenic Escherichia coli based on fourpatterns of virulence genes. , J Clin Microb 50, 1673-1678.
- 36.Mohamed L, Ge Z, Yuehua L, Yubin G, Rachid K et al.Virulence traits of avian pathogenic (APEC) and fecal (AFEC).
- 37.E. coli isolated from broiler chickens in Algeria. Tropical animal health and productionMar;50(3):. 547-53.
- 38.Unno T, Han D, Jang J, Widmer K, Ko G et al. (2011) Genotypic and phenotypic trends in antibiotic resistant pathogenic Escherichia coli isolated from humans and farm animals in South Korea. Microbes and environments. 26(3), 198-204.
- 39.Johnson T J, Wannemuehler Y, Doetkott C, Johnson S J, Rosenberger S C et al. (2008) Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. , J Clin Microb 46, 3987-3996.
- 40.M J Schwaber, Carmeli Y. (2008) Carbapenem-resistant Enterobacteriaceae: a potential threat. , Jama 300(24), 2911-2913.
- 41.Nordmann P, Naas T, Poirel L. (2011) Global spread of carbapenemase-producing Enterobacteriaceae. Emerging infectious diseases. 17(10), 1791.
- 42.WHO. (2017) Health Organization), “Critically important antimicrobials list. 5th rev,”. http://who.int/foodsafety/publications/antimicrobials-fifth/en/..
- 43.Fang D O N G, L U Jie, Yan W A N G, Jin S H I, J H Zhen et al. (2017) A five-year surveillance of carbapenemase-producing Klebsiella pneumoniae in a pediatric hospital in China reveals increased predominance of NDM-1. Biomedical and environmental sciences. 30(8), 562-569.
- 44.K, M A Toleman, T R Walsh, Bagaria J, Butt F et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. The Lancet infectious diseases. 10(9), 597-602.
- 45.W J Liang, Liu 43Ahmed, M A, Shimamoto T, Shimamoto T. (2013) Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. , International Journal of Medical Microbiology 303-8.
- 46.Yong D, M A Toleman, C G Giske, H S Cho, Sundman K et al. (2009) Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrobial agents and chemotherapy. 53(12), 5046-5054.
- 47.T R Walsh. (2010) Emerging carbapenemases: a global perspective. , International journal of antimicrobial agents 36, 8-14.
- 48.D M Livermore. (2009) Has the era of untreatable infections arrived?. , Journal of Antimicrobial Chemotherapy 64, 29-36.
- 49.Ahmad K, Khattak F, Ali A. (2018) Carbapenemases and extended-spectrum β-lactamase–producing multidrug-resistant Escherichia coli isolated from retail chicken in Peshawar: first report from Pakistan,”. , Journal of Food Protection 81, 1339-1345.
- 50.Poirel L, Berçot B, Millemann Y, R A Bonnin, Pannaux G et al. (2012) Carbapenemase-producing Acinetobacter spp. in cattle. , France,” Emerging Infectious Diseases 18, 523-525.
- 51.P L Ho, Y, Wang Y, W U Lo, Lai E L Y et al. (2016) Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. , European Journal of Clinical Microbiology & Infectious Diseases 35, 379-385.
- 52.Tian X, Zheng X, Sun Y, Fang R, Zhang S et al. (2020) Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002–2017. Infection and Drug Resistance. 501-512.
- 53.Gülmez D, Woodford N, Palepou M F I, Mushtaq S, Metan G et al. (2008) Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. International journal of antimicrobial agents. 31(6), 523-526.
- 54.V C Cheng, S C Wong, P L Ho, K Y Yuen. (2015) Strategic measures for the control of surging antimicrobial resistance in Hong Kong and mainland of China. Emerging microbes & infections. 4-1.
